AOP networks¶
Effects are related to each other using the toxicological concept of adverse outcome pathways (AOPs) and adverse outcome pathway networks (see https://aopwiki.org). Adverse Outcome Pathway (AOP) Networks specify how biological events (effects) can lead to an adverse outcome (AO) in a qualitative way through relations of upstream and downstream key events (KEs), starting from molecular initiating events (MIEs). Using AOPs, the adverse outcome (AO), e.g., liver steatosis, is linked to key events (KEs), e.g., triglyceride accumulation in the liver, and to molecular initiating events (MIEs), e.g., PPAR-alpha receptor antagonism. In general, multiple AOPs may lead to the same AO, and therefore AOP networks can be identified.
This module has as primary entities: Effects
Output of this module is used by: QSAR membership models Molecular docking models Active substances Relative potency factors Hazard characterisations Points of departure Effect representations
AOP networks as data¶
Settings and Tiers¶
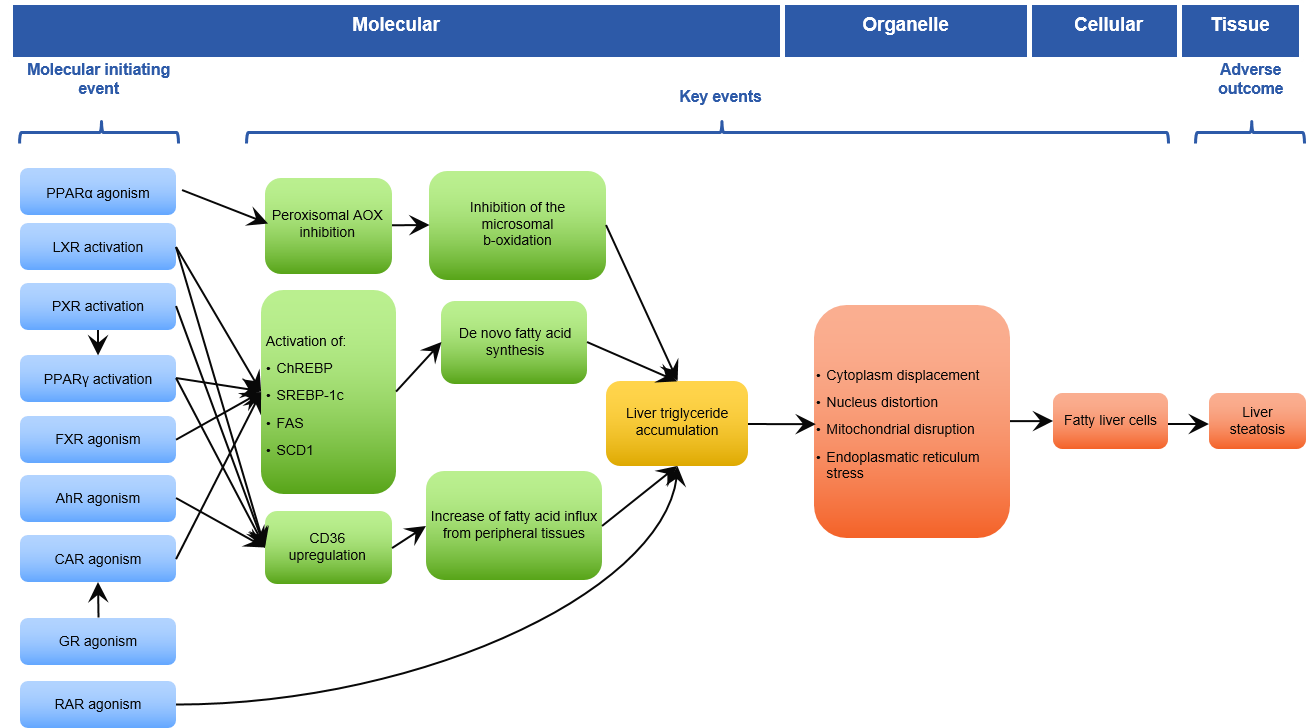
Figure 51 AOP network¶